
Besides, the ultra-long noisy reads with an average length over 50-Kb from Oxford Nanopore Technology (ONT) and related software development such as wtdbg2, also largely improve the contig continuity in genome assemblies. Specific software for assembling HiFi reads have also been developed, such as Hifiasm and HiCanu, which in general can produce large contigs with N50 size over tens of mega-bases for a substantial part of genomes. The long accurate reads generated by PacBio High-Fidelity (HiFi) technology, with an average read length over 10-Kb and base accuracy over 99%, has been widely applied in the de novo assembly of plant and animal genomes.
Chromosomal scaffold meaning manual#
As more genome projects have been launched and the contig continuity constantly improved, we believe EndHiC has the potential to make a great contribution to the genomics field and liberate the scientists from labor-intensive manual curation works. EndHiC is efficient both in time and memory, and it is interface-friendly to the users. ConclusionsĮndHiC is a novel Hi-C scaffolding tool, which is suitable for scaffolding of contig assemblies with contig N50 size near or over 10 Mb and N90 size near or over 1 Mb. EndHiC has been successfully applied in the Hi-C scaffolding of simulated data from human, rice and Arabidopsis, and real data from human, great burdock, water spinach, chicory, endive, yacon, and Ipomoea cairica, suggesting that EndHiC can be applied to a broad range of plant and animal genomes. Benefiting from the increased signal to noise ratio, the reciprocal best requirement, as well as the robustness evaluation, EndHiC achieves higher accuracy for scaffolding large contigs compared to existing tools.
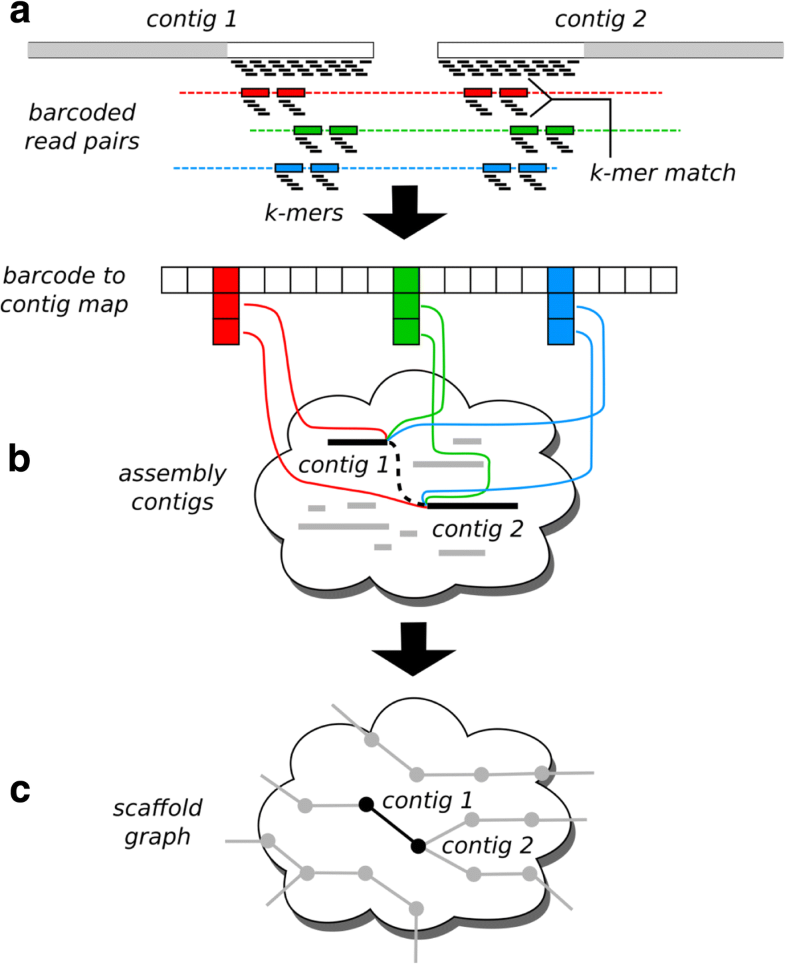
By this way, the signal neighbor contig linkages and noise non-neighbor contig linkages are separated more clearly. The core idea behind EndHiC, which distinguishes it from other Hi-C scaffolding tools, is using Hi-C links only from the most effective regions of contig ends. We design and develop a novel Hi-C based scaffolding tool EndHiC, which is suitable to assemble large contigs into chromosomal-level scaffolds. As the Hi-C links of two adjacent contigs concentrate only at the neighbor ends of the contigs, larger contig size will reduce the power to differentiate adjacent (signal) and non-adjacent (noise) contig linkages, leading to a higher rate of mis-assembly. The application of PacBio HiFi and ultra-long ONT reads have enabled huge progress in the contig-level assembly, but it is still challenging to assemble large contigs into chromosomes with available Hi-C scaffolding tools, which count Hi-C links between contigs using the whole or a large part of contig regions.


 0 kommentar(er)
0 kommentar(er)
